Paper Review: BiomedGPT: A Unified and Generalist Biomedical Generative Pre-trained Transformer for Vision, Language, and Multimodal Tasks
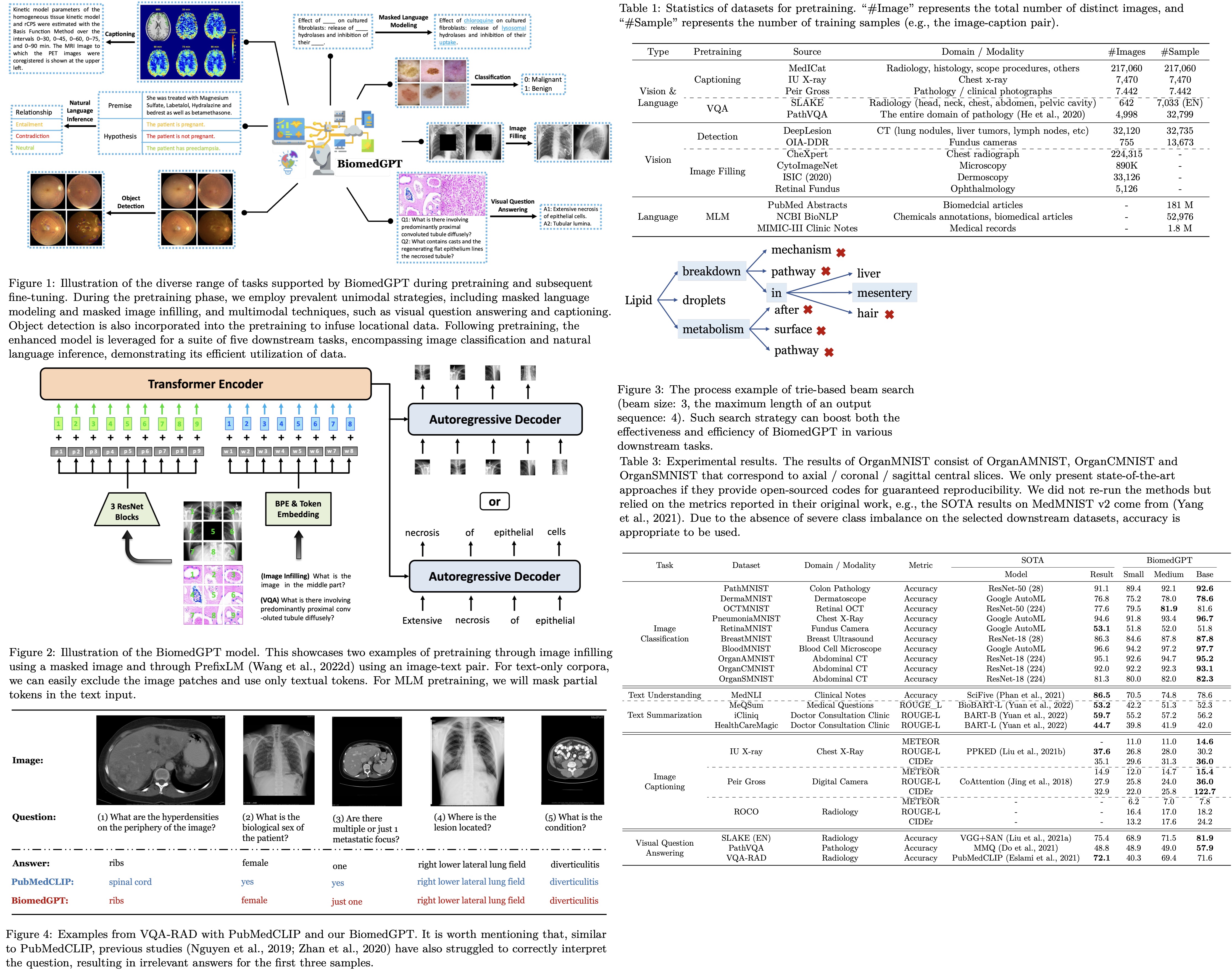
The paper introduces a new model called BiomedGPT (Biomedical Generative Pre-trained Transformer). BiomedGPT is designed to handle various types of biomedical data and perform a range of tasks by using self-supervision on large and diverse datasets. The experiments conducted show that BiomedGPT outperforms most previous state-of-the-art models across five different tasks and 20 public datasets, covering 15 unique biomedical modalities. The authors also demonstrate the effectiveness of their multi-modal and multitask pretraining approach in transferring knowledge to new and unseen data. The paper represents a significant advancement in developing unified and versatile models for biomedicine, with the potential to improve healthcare outcomes.
BiomedGPT Pipeline
Architecture selection
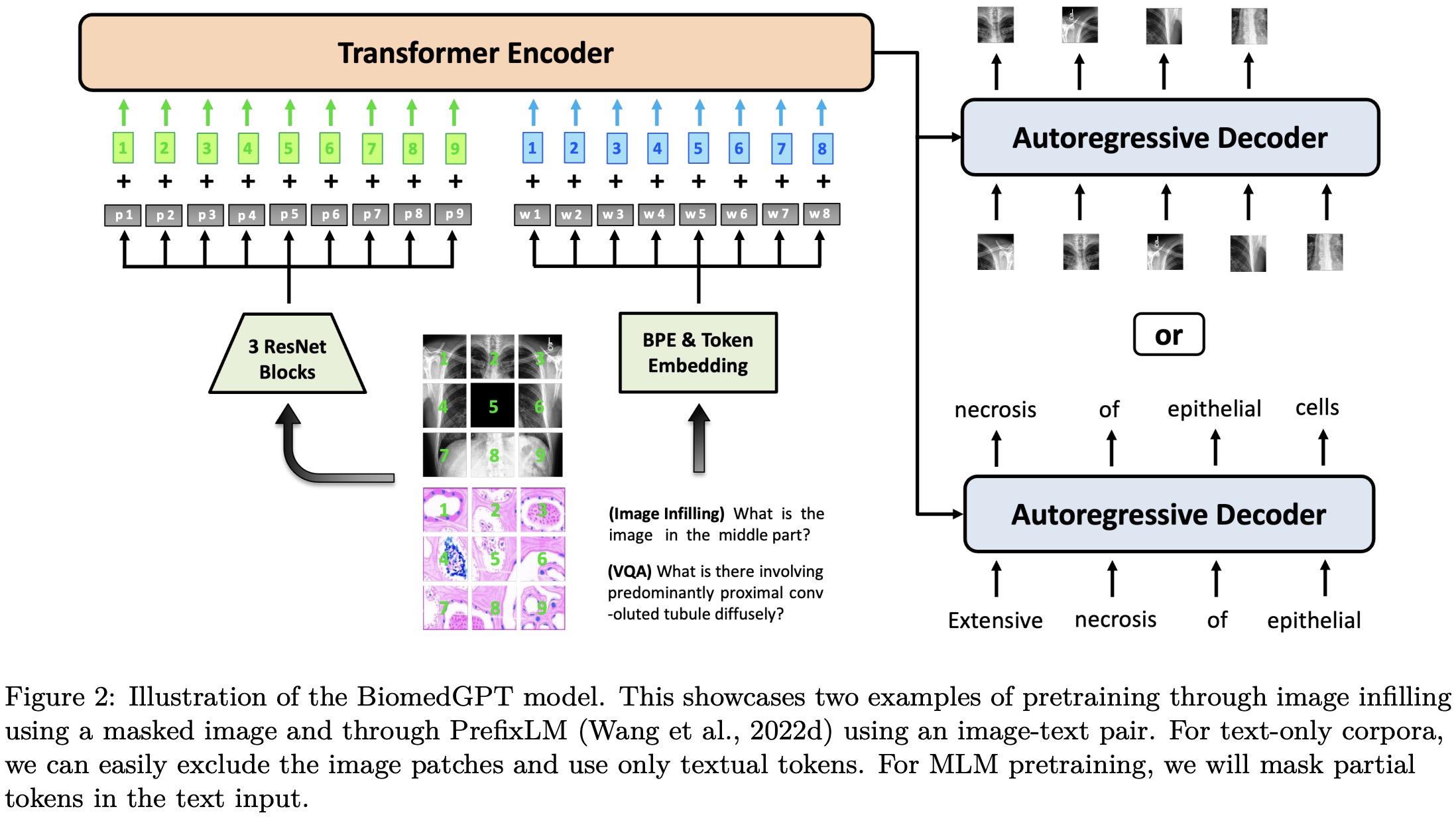
The proposed BiomedGPT model adopts an encoder-decoder architecture, which effectively maps various modalities into a unified semantic representation space and handles diverse tasks.
BiomedGPT is designed based on the BART architecture, a sequence-to-sequence model with a BERT-style encoder and a GPT-style decoder. The authors make architectural changes to improve convergence efficiency and stability in pretraining: they add normalization operations, incorporate absolute and relative position embeddings for text and images, and implement decoupling to separate position correlation. These modifications enhance the model’s ability to encode positional information and handle different modalities.
Input/Output Unification
BiomedGPT model processes inputs with various modalities, including images, language, and bounding boxes. To achieve a unified representation, the model applies CNN backbones to extract image features and ResNet modules to map images into a sequence of patches. For linguistic inputs, byte-pair encoding (BPE) is used for subword tokenization.
To handle diverse modalities without relying on task-specific output structures, the model represents them with tokens from a finite vocabulary. Frozen image quantization and object descriptor techniques are used to discretize images and objects. Text outputs, such as object labels and summarizations, are represented with BPE tokens. The image is encoded into a sparse sequence to reduce its representation length, and bounding boxes are expressed as sequences of location tokens.
Natural Language as a Task Manager
Inspired by previous literature on prompt/instruction learning, the model specifies each task with a handcrafted instruction to eliminate task-specific modules. BiomedGPT supports various task abstractions, including vision-only, text-only, and vision-language tasks.
During pretraining, the model performs tasks such as masked image modeling (MIM) and image infilling, where it recovers masked patches in images by generating corresponding codes (What is the image in the middle part?). It also learns object detection by generating bounding boxes of objects in images (What are the objects in the image?). For the text-only task, the model utilizes masked language modeling (MLM) where it predicts masked words in sentences (What is the complete text of ‘A case of oral
During fine-tuning and inference, the model extends its capabilities to tasks beyond pretraining, including image classification, text summarization, and natural language inference. Specific instructions are provided for each task to guide the model’s behavior and predictions.
Generative Pretraining via Seq2seq
Autoregressive or sequence-to-sequence (seq2seq) modeling is widely used in sequential tasks, including large language modeling. In the case of BiomedGPT, the model’s architecture is parametrized by θ, and it is trained autoregressively using the chain rule.
BiomedGPT incorporates both linguistic and visual tokens in its input sequence, such as subwords, image codes, and location tokens. Subwords are obtained using a byte-pair encoding (BPE) tokenizer, and during the masked language modeling (MLM) task, 30% of the subword tokens are masked. For the object detection (OD) task, location tokens are generated using Pix2Seq conditioned on observed pixel inputs. Biomedical images undergo preprocessing using VQ-GAN for quantization, involving removal of trivial backgrounds, cropping to the object of interest, resizing, and generating sparse image codes as the target output for the masked image modeling (MIM) task. Vision-language tasks follow a similar tokenization process.
During fine-tuning, the model continues to utilize seq2seq learning, but with different datasets and tasks.
Autoregressive Inference
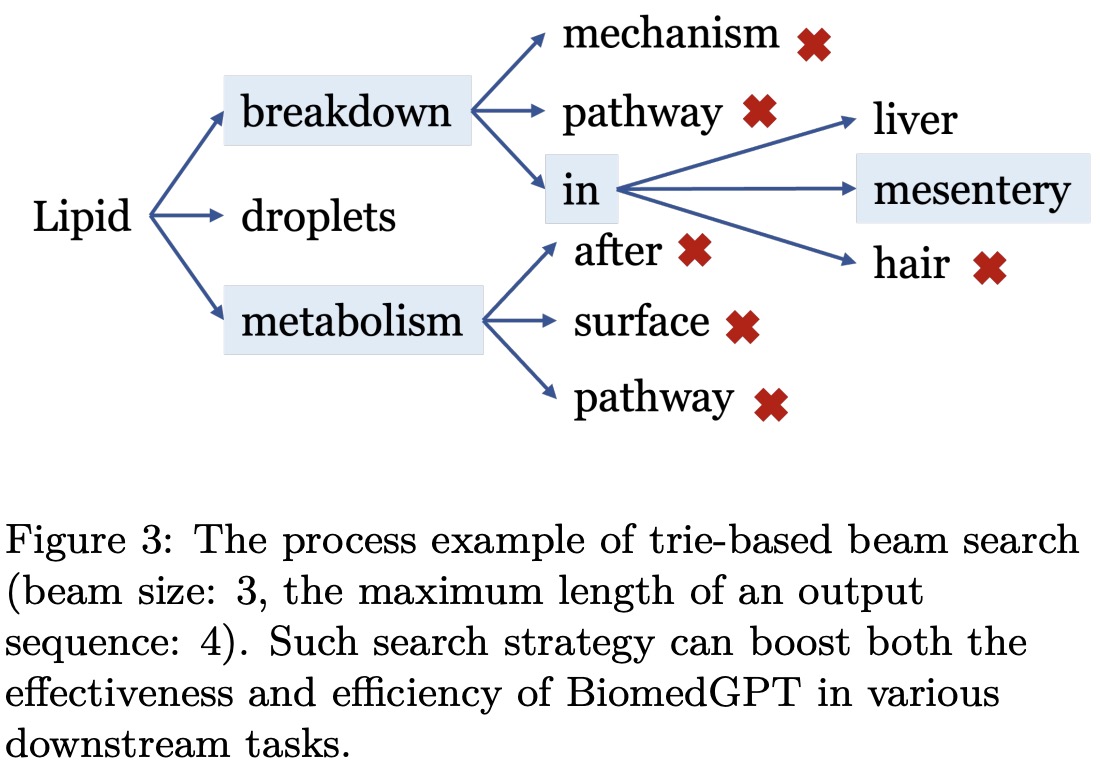
To enhance generation quality and address challenges in classification tasks, such as unnecessary optimization of the entire vocabulary and the generation of invalid labels, a beam search strategy incorporating a prefix tree (trie) is applied. This strategy limits the number of candidate tokens and improves the efficiency and accuracy of decoding. During trie-based beam search, BiomedGPT sets logits for invalid tokens to -∞, ensuring that only valid tokens are considered. The trie-based search approach is also employed during the validation phase of the fine-tuning stage to accelerate the process, resulting in a significant increase in speed according to experiments conducted by the authors.
Experiments
The models are pre-trained with 10 Nvidia A5000 GPUs and mixed precision, except for the models used for fine-tuning downstream tasks.
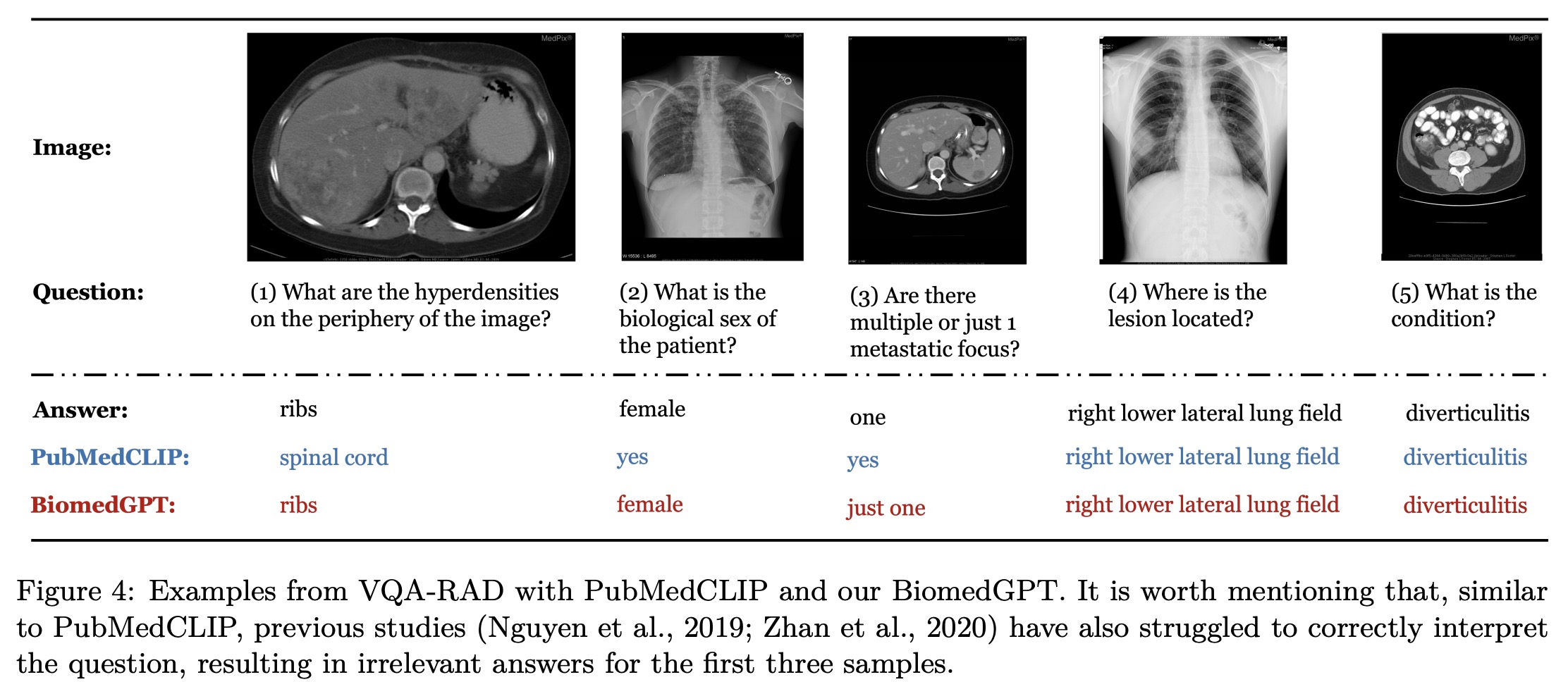
Results on Unimodal Datasets
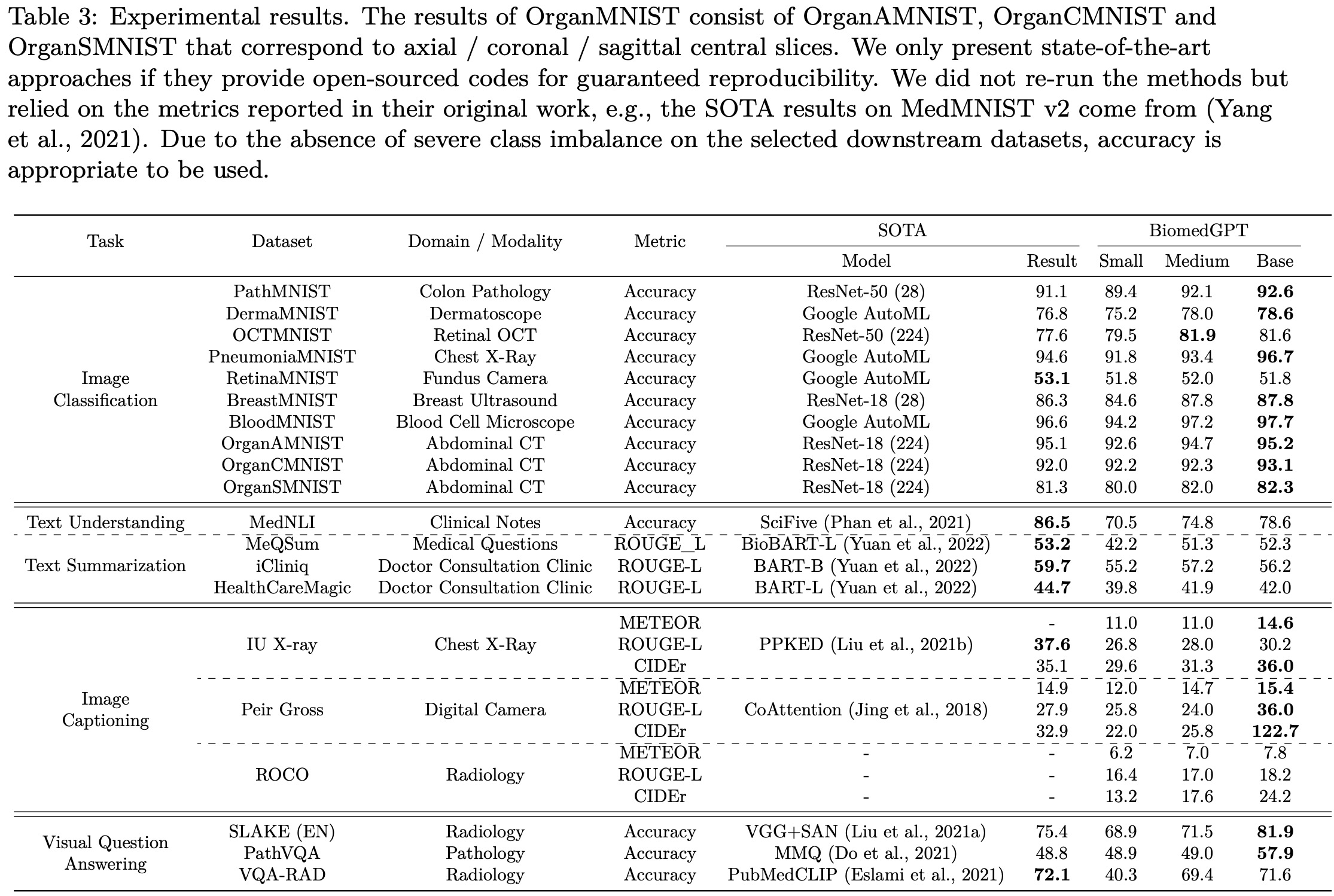
The authors conducted experiments on three unimodal tasks using 14 datasets. For the image classification task on MedMNIST v2, their BiomedGPTBase model achieved state-of-the-art accuracy on 9 out of 10 image-only datasets, with a small performance gap observed on RetinaMNIST, which was originally designed for regression.
In text-only tasks, their model achieved an accuracy of 78.6% on the natural language inference task using the MedNLI dataset, which is lower than the state-of-the-art result of 86.5%. For text summarization of doctor-patient conversations, their model did not achieve satisfactory performance as measured by the ROUGE-L metric. Several potential reasons were identified for the performance difference compared to previous state-of-the-art models: the constrained scale of their model with fewer parameters, a smaller corpus used for training compared to other models, and the impact of divergent data between biomedical articles and clinical notes.
Results on Multimodal Datasets
BiomedGPT focuses on two multimodal tasks: image captioning and visual question answering (VQA). They select specific datasets for each task, including SLAKE, PathVQA, VQA-RAD, IU X-ray, PEIR Gross, and ROCO. Due to limited availability of public multimodal biomedical datasets, the model is pre-trained on training sets and evaluated on unseen testing sets.
BiomedGPT outperforms state-of-the-art methods in terms of CIDEr, particularly achieving a significant improvement of 273% on the PEIR Gross dataset. While falling short in terms of ROUGE-L on the IU X-ray dataset, the authors attribute this to their model selection based on CIDEr during the validation phase.
BiomedGPT achieves significant improvements over previous state-of-the-art models on the SLAKE (EN) and PathVQA datasets. They note that the performance improvement has not reached a plateau, and believe that increasing the number of fine-tuning epochs can further enhance accuracy. BiomedGPT generates semantically correct outputs in their case analysis, although the current evaluation framework may not fully reflect the model’s capability to produce accurate answers in terms of semantic understanding.
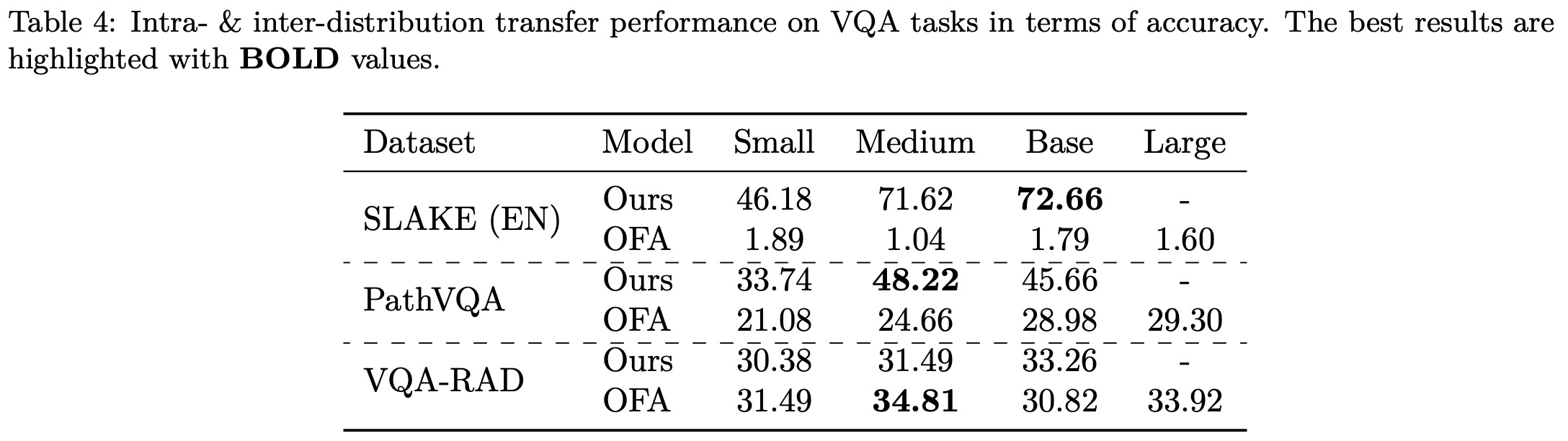
Results on Intra- & Inter-distribution Transfer
The authors conducted intra- and inter-distribution inference experiments to evaluate the performance of pre-trained BiomedGPT checkpoints on seen and unseen datasets. The checkpoints were pre-trained on SLAKE and PathVQA training sets and evaluated on the testing sets of those two datasets and VQA-RAD. Results show BiomedGPT achieves excellent performance, especially with larger models, on SLAKE and PathVQA. In contrast, the baseline OFA model, pre-trained on general datasets like ImageNet, experiences significant performance degradation on these two datasets.
The authors hypothesize that the models may overfit the familiar intra-domain distribution and struggle with out-of-distribution data. They observe that BiomedGPT tends to overfit intra-distribution instructions, leading to difficulties in understanding unseen questions and generating uncontrolled outputs. They provide an example where BiomedGPTBase mistakenly interprets a VQA task as an image generation task. Although models generate text-only answers that match partially, they still do not match the open-ended ground truth. The models exhibit catastrophic performance on open-ended questions in the VQA-RAD test set.
These observations highlight the challenge of instruction-sensitivity in instruction-guided pretraining when developing a unified and generalist biomedical model. The authors plan to explore data augmentation and synthetic biomedical datasets in future work to address the limited volume and diversity of existing biomedical datasets, as well as concerns regarding data privacy and imbalance.
Discussion
Main Findings:
- BiomedGPT achieves competitive performance across various tasks in vision, language, and multimodal domains by integrating diverse biomedical modalities and tasks within a unified seq2seq pretraining framework.
- Including a broad spectrum of biomedical tasks and modalities in pretraining significantly enhances fine-tuning efficiency and overall model performance.
- The observation that OFA, a generalist model pre-trained with generic data, struggles to align image-text pairs during fine-tuning highlights the importance of effective multi-modal, multi-task pretraining, which BiomedGPT addresses.
- Scaling up the model size leads to a considerable boost in performance.
Limitations and Suggestions:
- BiomedGPT shows sensitivity to instructions, sometimes failing to understand instructions and making catastrophic predictions. Increasing the diversity of high-quality instruction sets during pretraining could be a solution.
- Balancing data diversity, such as size ratios and training sequences for different biomedical modalities, needs to be explored.
- Reinforcement learning from human or AI feedback (RLF) could be employed to align BiomedGPT with human intent, but creating specific biomedical RLF datasets would be expensive.
- The considerable difference between clinical notes and general/biomedical text, as well as the presence of vision-only tasks, pose challenges for text-only downstream tasks. Generating a representative vocabulary from all domains and adjusting the ratio of text inputs during pretraining could help address these issues.
- Fine-tuning efficiency, in terms of training speed and memory bottleneck, is a concern for large-scale generalist biomedical models. Parameter-efficient fine-tuning (PEFT) is a potential research direction to explore.
- The attempt to apply prompt tuning did not yield expected results.