Paper Review: UniverSeg: Universal Medical Image Segmentation
UniverSeg is a new method for medical image segmentation that can generalize to unseen tasks, which involves new anatomies, image modalities, or labels, without needing additional training. This is a solution to the problem of time-consuming model training or fine-tuning typically required by researchers when faced with a new segmentation task. UniverSeg uses a Cross-Block mechanism to create accurate segmentation maps given a query image and a set of image-label pairs that define the new task. To ensure its ability to generalize, UniverSeg was trained on MegaMedical, a vast collection of 53 open-access medical segmentation datasets with over 22,000 scans. The method was found to outperform several other approaches on unseen tasks, providing useful insights into the proposed system’s functioning.
UniverSeg Method
Traditional segmentation strategies utilize parametric functions that are usually modeled using a convolutional neural network. These functions learn to predict segmentation for a specific task, based on a set of image-label pairs associated with that task. However, this approach limits the model’s capability to only that particular task. In contrast, the proposed method learns a universal function that can predict a label map for any given task, according to the support (set of example image-label pairs) available for that task. This allows the model to generalize to new, unseen tasks.
Model
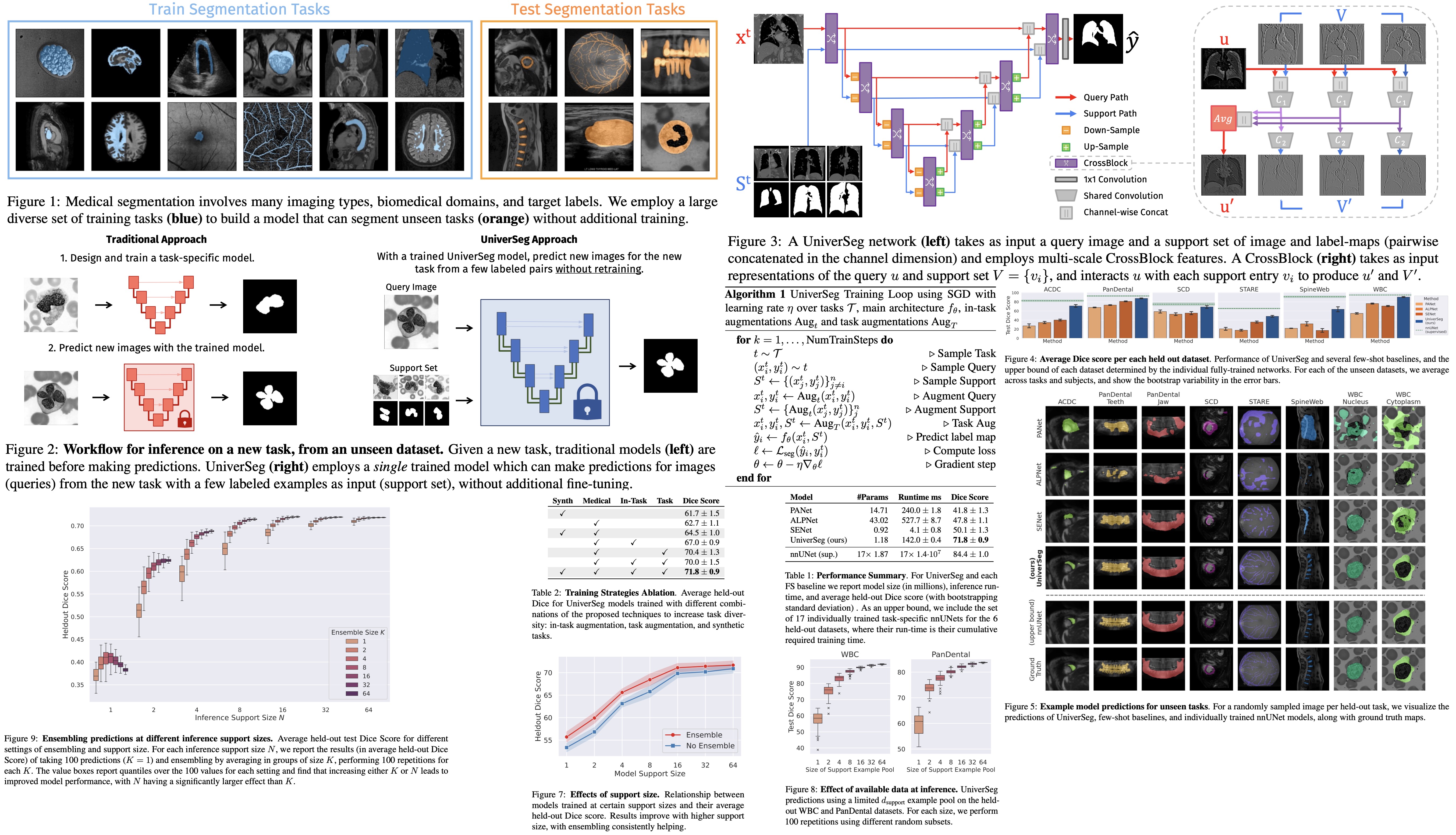
The authors implement a universal function using a fully convolutional neural network that combines a newly proposed CrossBlock module and cross-convolution layer. The CrossBlock module employs a cross-convolution layer to transfer information between the support set and the query image. This layer performs permutation-invariant operations due to weight reuse. The CrossBlock module produces updated query representations and supports at each network step.
The neural network uses an encoder-decoder structure with residual connections similar to the popular UNet architecture to integrate information across spatial scales. The network inputs include a query image and a support set consisting of image and label map pairs, each concatenated channel-wise. The output is a segmentation prediction map.
Each level in the encoder path includes a CrossBlock followed by a spatial down-sampling operation on both query and support set representations. Each level in the expansive path consists of up-sampling both representations, doubling their spatial resolution, concatenating them with the equivalently-sized representation in the encoding path, followed by a CrossBlock. Finally, a single 1x1 convolution is performed to map the final query representation to a prediction.
Training
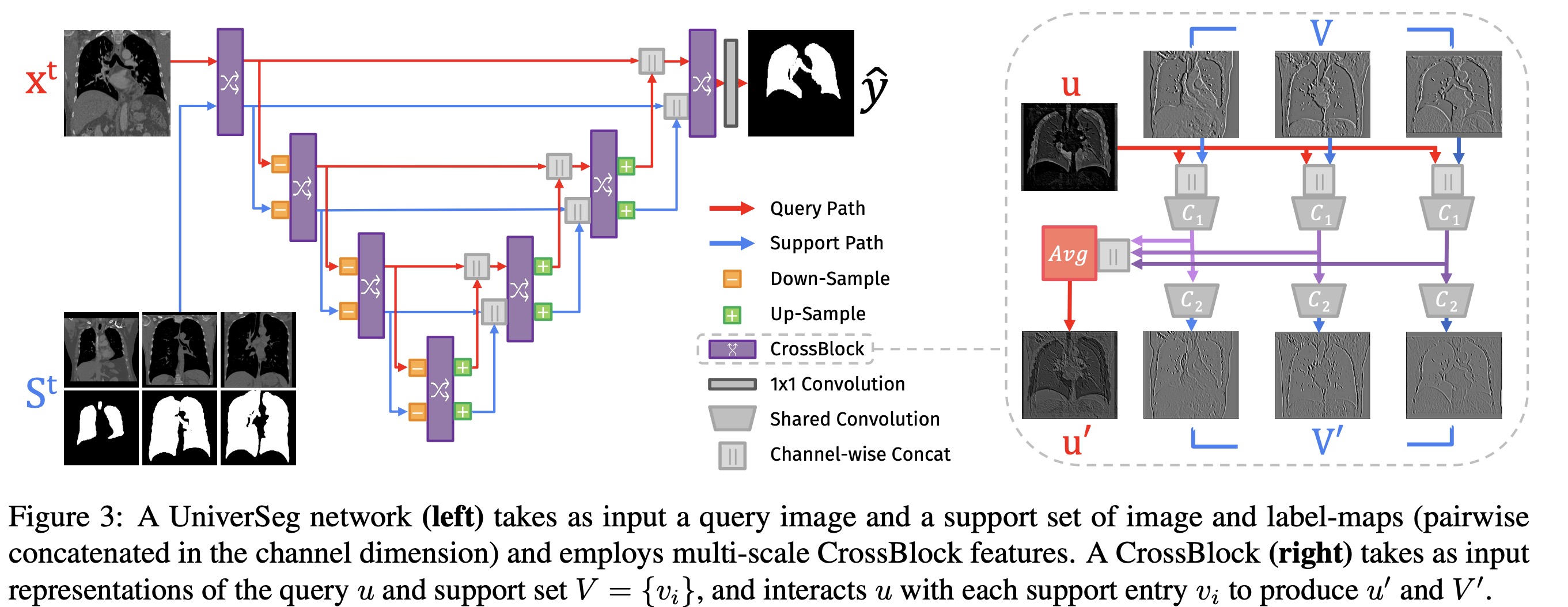
UniverSeg is trained using a wide variety of training tasks. The loss function is a standard segmentation loss such as cross-entropy or soft Dice, which measures the agreement between the predicted and ground truth values. To increase the variety of training tasks and effective training examples, the researchers employed data augmentation strategies.
- In-Task Augmentation reduces overfitting to individual subjects by applying standard data augmentation operations like affine transformations, elastic deformation, or adding image noise to the query image and each entry of the support set independently.
- Task Augmentation helps the model generalize to new tasks, particularly those far from the training task distribution. Task augmentation involves altering all query and support images and/or all segmentation maps with the same type of task-changing transformation. Examples of task augmentations include edge detection of segmentation maps or applying a horizontal flip to all images and labels.
MegaMedical Dataset
To train the universal model, UniverSeg, a large and diverse set of segmentation tasks was employed, collected in a dataset called MegaMedical. MegaMedical is an extensive collection of 53 open-access medical segmentation datasets, covering 26 medical domains and 16 imaging modalities.
The datasets were standardized across different formats and expanded using synthetic segmentation tasks to further increase diversity. MegaMedical covers a broad array of biomedical domains such as eyes, lungs, spine vertebrae, white blood cells, abdomen, and brain. The datasets vary in acquisition details, subject age ranges, and health conditions.
While the datasets in MegaMedical offer a variety of imaging tasks and label protocols, the research primarily focuses on 2D binary segmentation. For datasets with 3D data, 2D mid-slices were extracted, and for those with multiple segmentation labels or modalities, multiple tasks were created. All images were resized to 128x128 pixels and intensity normalized to the range [0,1].
The researchers also adapted a procedure to generate a thousand synthetic tasks using random synthetic shapes, which were used alongside the medical tasks during training.
Experiments
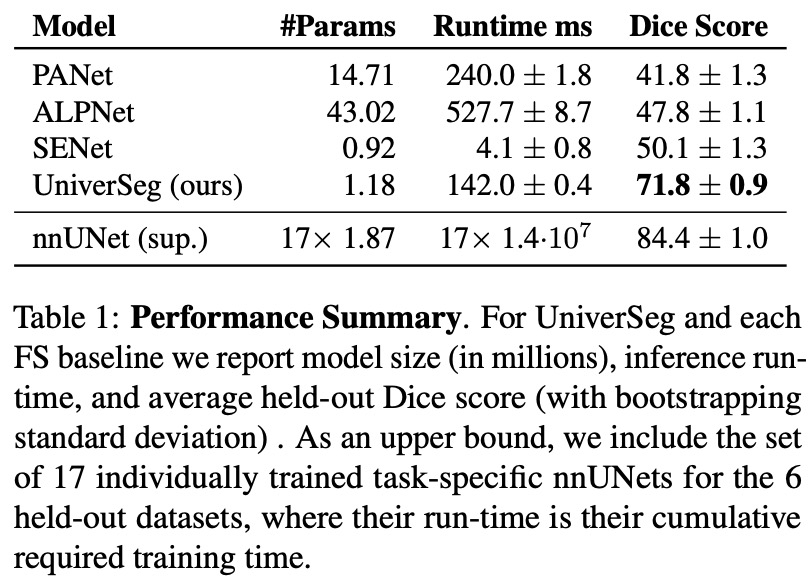
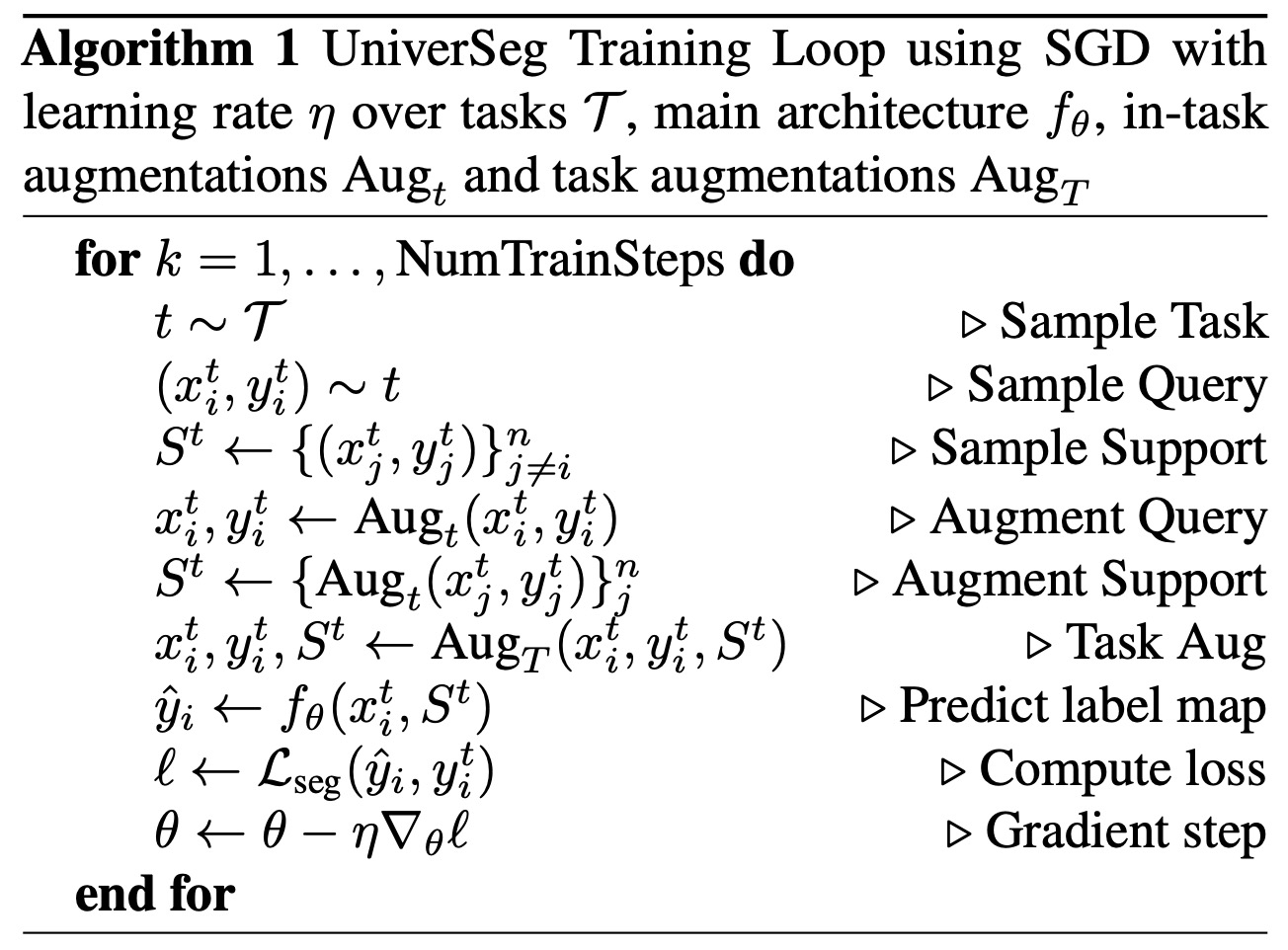
- The authors compared the segmentation quality of UniverSeg with Few-Shot baselines and task-specific upper bounds to assess UniverSeg’s effectiveness in solving tasks from unseen datasets. Results show that UniverSeg significantly outperforms all FS methods in all held-out datasets.
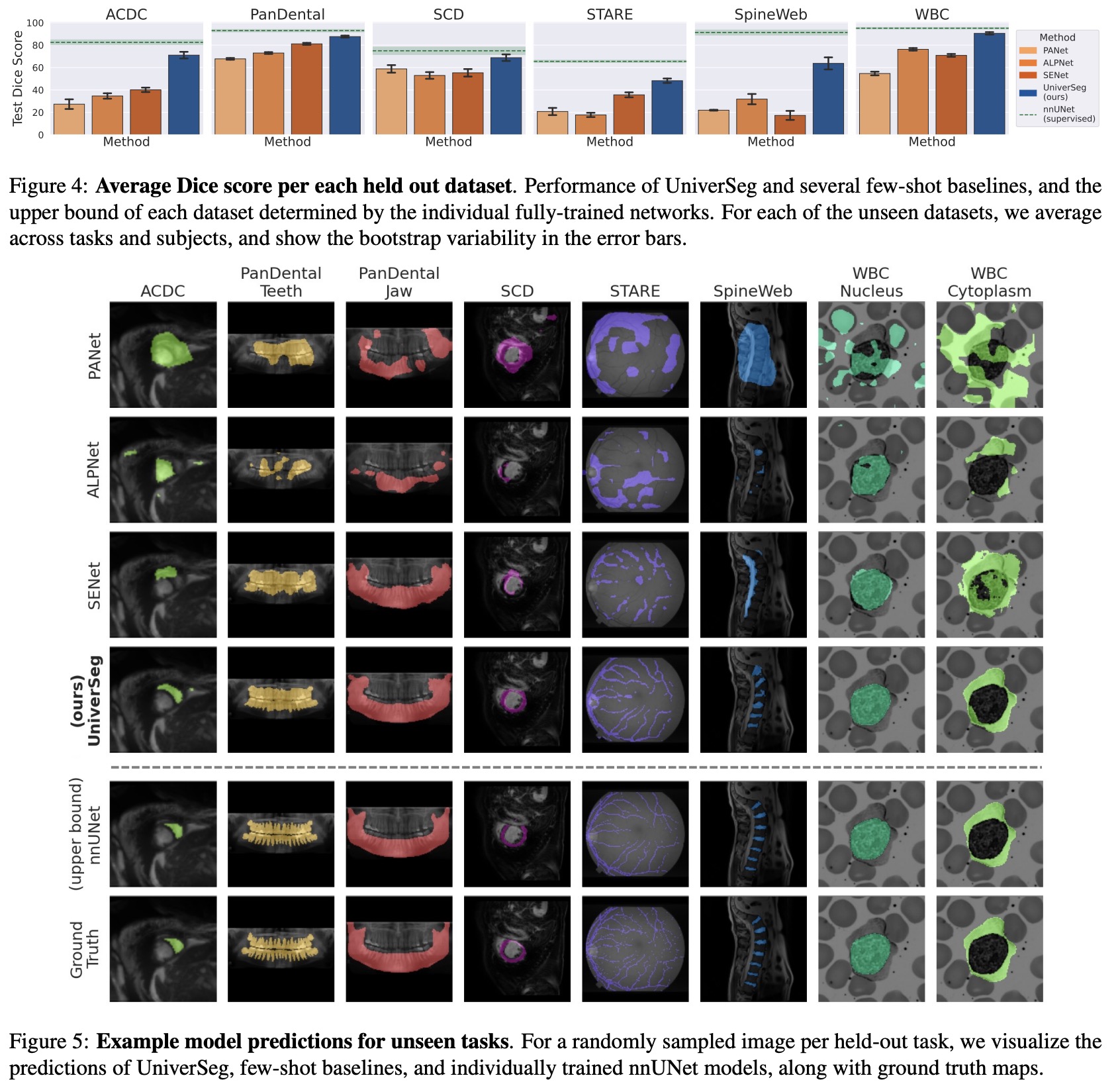
- Improvements in Dice scores range from 7.3 to 34.9 when comparing UniverSeg with the highest performing baseline for all datasets. The visual representation of the predicted segmentations also shows clear qualitative improvements.
- Notably, UniverSeg requires fewer model parameters than other models like PANet, ALPNet, and the nnUNets, and a similar number to SENet. Despite the similarities between SENet and UniverSeg, the results suggest that the proposed CrossBlock in UniverSeg is better suited to transferring spatial information from the support set to the query.
- Interestingly, for some datasets such as PanDental or WBC, UniverSeg performs competitively with supervised task-specific networks that were extensively trained on each of the held-out tasks. The results also show that segmentations produced by UniverSeg more closely match those of the supervised baselines than those of any other few-shot segmentation task, especially in challenging datasets like SpineWeb or STARE.
Ablations
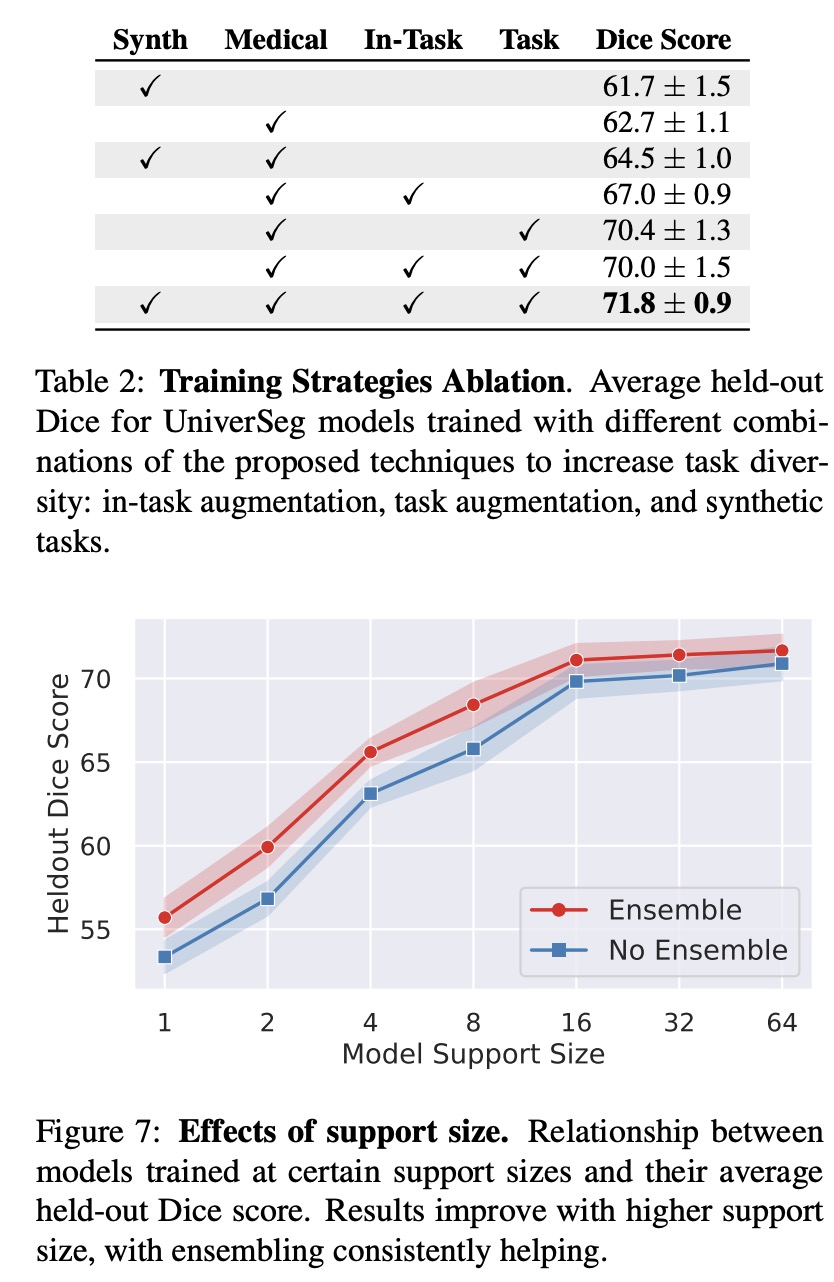
- Task Quantity and Diversity: More training tasks improve performance on held-out tasks. However, the choice of datasets can also significantly impact the results. For instance, broader anatomical diversity in training datasets led to better model performance.
- Ablation of Training Strategies: All proposed strategies (in-task augmentation, task augmentation, synthetic tasks) increased model performance, with the best results achieved when using all strategies jointly. Interestingly, a model trained only on synthetic data also performed well on medical tasks despite not being exposed to medical training data, underscoring the importance of task diversity.
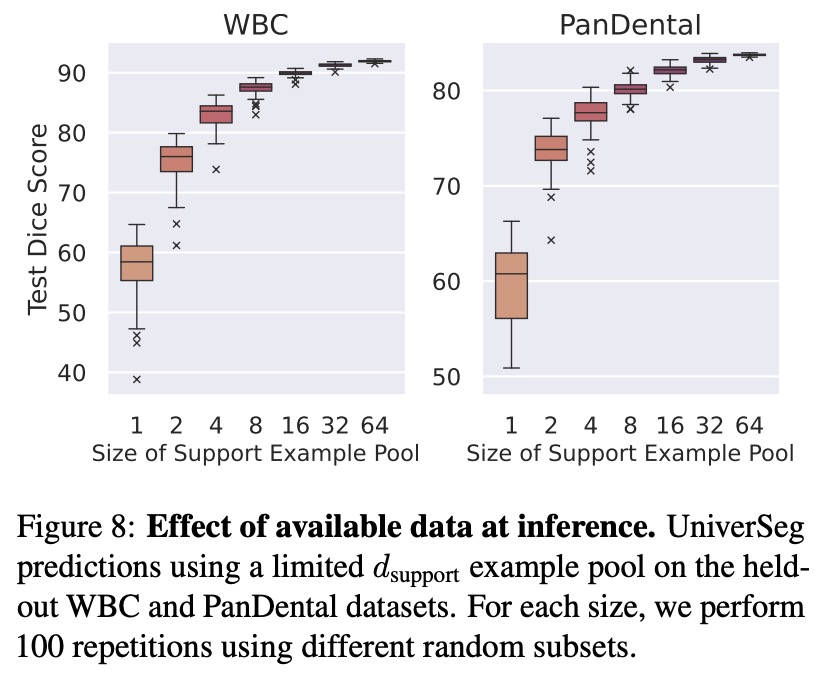
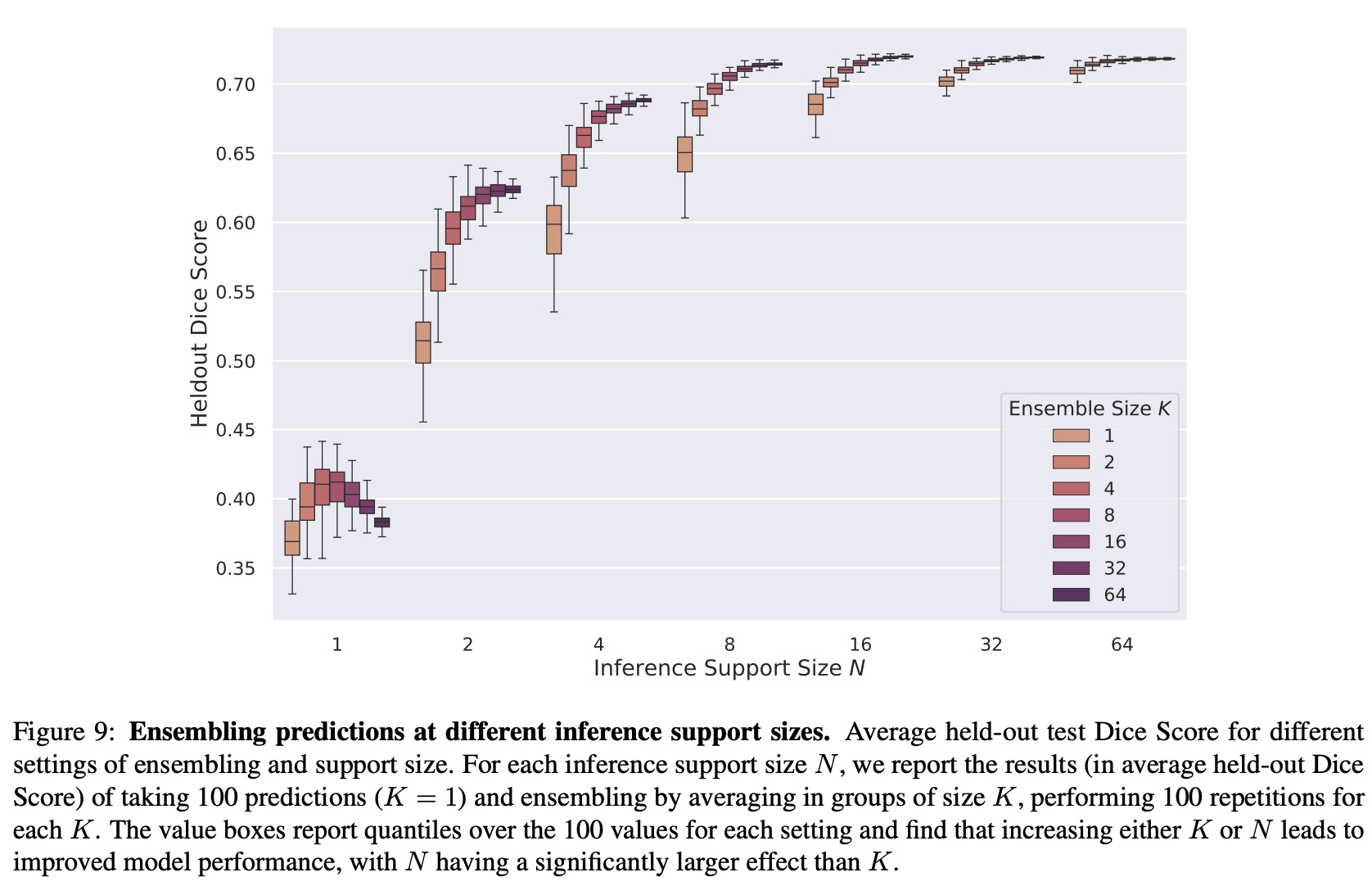
- Support Set Size: Best results were achieved with larger training support set sizes, with diminishing returns beyond a certain size. Ensembling predictions consistently improved results, especially for smaller support sets.
- Limited Example Data: The number of labeled images significantly affects UniverSeg performance. As the support size increased, both average segmentation quality and stability improved due to the reduction of variance from diverse support sets.
- Support Set Ensembling: Increasing both support size and ensemble size improved results and reduced variance. However, increasing the support size had a more significant impact than increasing the ensemble size. This suggests that UniverSeg models exploit information from the support examples in a different way than traditional ensembling techniques in few-shot learning.