How we tried to create a medical chatbot. The story of a project that never got the green-light
In recent years, the telemedicine market (remote medical services) and MedTech, in general, have been snowballing, and the coronavirus pandemic has only accelerated its development. Such technologies are in demand because they are relatively cheap, available regardless of where the patient lives, and allow the patients to choose their own doctors.
However, there are many problems in implementing and delivering these technologies, such as slow legislation changes and difficulties in obtaining, processing and storing confidential data. Currently, in Russia, doctors are not allowed to make a diagnosis without meeting with patients face-to-face. Meanwhile, according to an order from the Ministry of Medicine, by 2030, half of the medical consultations should take place online.
Online consultations are very popular in MedTech nowadays. They can be done with or without a doctor. Naturally, serious issues should be solved by a doctor, but there are still many simple tasks that artificial intelligence can handle. Such solutions have already been implemented in Russia: bots help to make appointments, conduct surveys before these appointments, analyze doctor-patient dialogues, and convert audio recordings into text.
At MTS AI, we had been developing our medical chatbot for about two years, and… it didn’t work out. The idea was simple: before the first appointment, the patient speaks with the chatbot, which conducts an initial survey. The output is an anamnesis for the doctor, and the patient gets to know which specialist he should go to. The benefit for the doctor is that he does not have to ask the same questions to each patient but needs only to verify the anamnesis and the presumed diagnosis. This reduces the initial appointment’s time length and speeds up patient flow. In addition, the patient does not always know which doctor to go to, and the chatbot will help him navigate. Of course, it would be better if the chatbot could show the potential diagnosis, but this is risky and could violate the law, so we decided to give only recommendations.
The project turned out to be very complicated, its potential profitability was unclear, so it was stopped at the end of 2020. But we learned a lot while working on it, and I’d like to share that.
I’ve been a tech lead of the NLP part of the project for about a year, and I’m going to talk about that.
In this article, I will describe the work on the project as a whole first, and then I will speak on data collection and labeling. As we know, the project’s success directly depends on the data’s quality. That is why it is essential to organize not only the data collection and labeling but also the validation of this labeling. After that, we’ll look at what machine learning models have been used - that’s the fun part, right? Then I’ll talk about which ideas didn’t work. And at the end, I will explain how we benefited from the project even though it did not work.
Additional notice: the project was initially in Russian (the original text can be found here, but I show the examples in English, so it would be easier to understand them.
High-level technical overview
Developing a high-quality chatbot is a big undertaking, and our project was no exception: data scientists in the field of NLP and recommendation systems, programmers, managers, and other specialists worked on it.
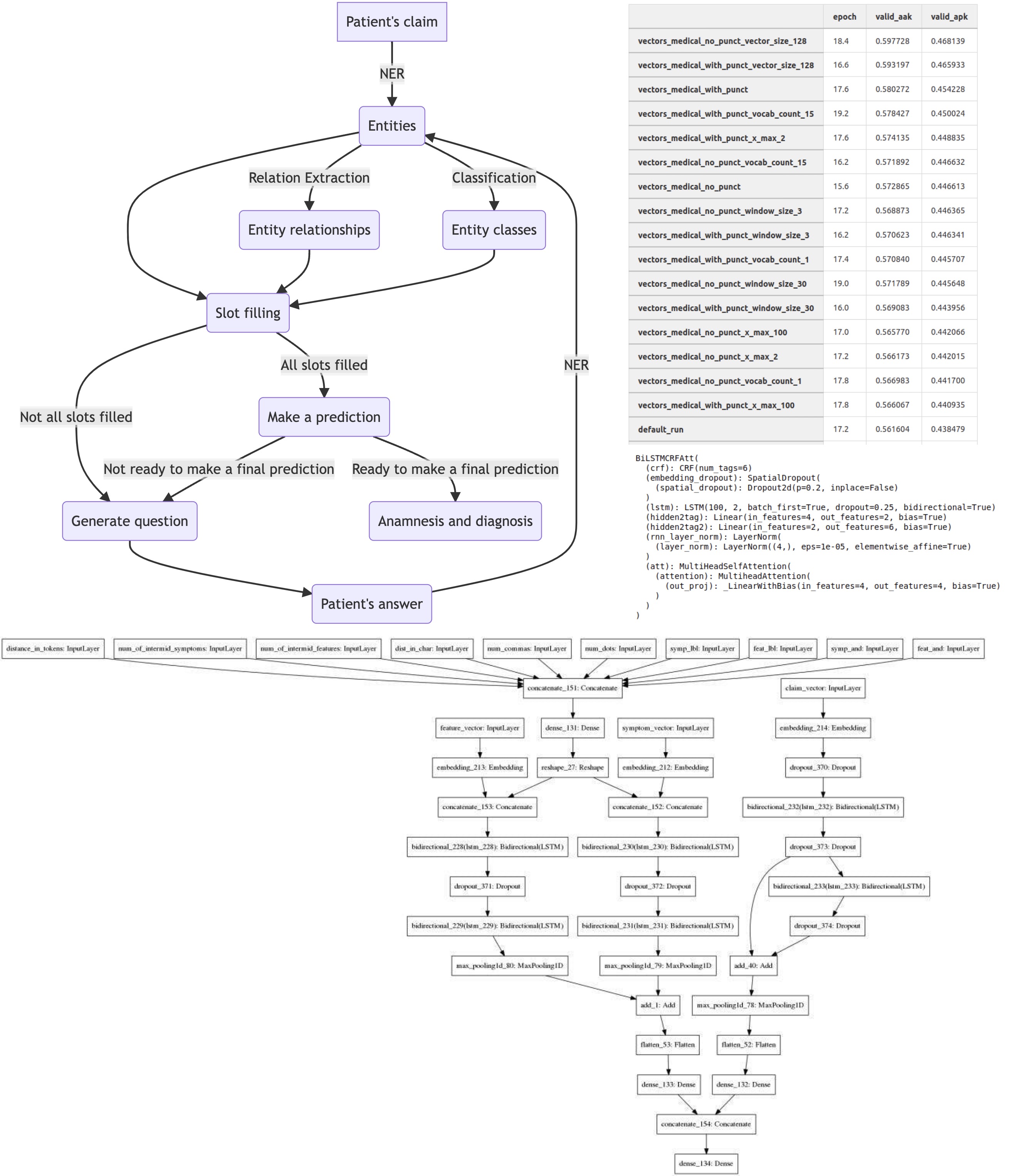
To begin with, let’s see how our chatbot should work. Here is a simplified scheme:
As mentioned above, the project performs two tasks: compiling an anamnesis and predicting a diagnosis (even if we won’t show it to the patient). To diagnose correctly, we need a detailed description of the person’s health. For example, if a patient complains about “sore throat,” “dry cough” and reports several other specific symptoms, we can say he probably has “acute pharyngitis.” But it is very difficult or impossible to extract such symptoms from the raw text, and it would take too long to ask people about all of them, as they would get tired of answering. So instead, we decided to extract separate entities from the text and combine them into more complex symptoms.
In the end, our team came up with the following scheme. We decided that patients would write unstructured text to the chatbot, and it was up to us to extract information from it and bring it into a structured form. This is what slot filling is usually used for.
We had a configuration file with lists of entities and links between them. We distinguished two groups: the entities themselves and their attributes. For example, an entity could be “pain”, “runny nose”, or “swelling”. Attributes can show under what conditions the pain occurs (condition), where it is felt (localization), when it occurs (time of day), etc. And it has been specified what attributes entities can have. Why is this necessary? Not all combinations make sense (for example, you don’t need to describe that a runny nose is felt in the nose - it doesn’t happen anywhere else), and some combinations are not important for diagnosis.
In addition, we had classes for most attributes. For example, it can hurt your arm, leg, left hypochondrium, ear, and other things. Symptoms can occur when sitting, bending over, breathing, etc. In addition, people may use diminutive suffixes, slang, and different alternative spellings. All of this makes it necessary to build classification models to predict specific classes.
Let’s see an example:
A patient writes us a complaint, such as: “I fell down the stairs, now my leg and arm hurt a lot, and my cheek also swells in the morning.” Note: The complaints here and hereafter are fictitious.

First, we extract entities using NER models. We need to extract a lot of them, so we use various approaches, from simple rule-based parsing and regular expressions to neural nets.
Next, we use the Relation Extraction model to find related entities. We need to figure out that the swelling occurs specifically in the morning and that only the leg and the arm hurt. To do this, we pass all pairs of entities and attributes and extra features through the model and make predictions. In addition, we set up filtering to discard impossible pairs.
The next step is splitting the attributes into classes (described above). This is a simple classification model for each term.
But that’s not all: the complaint could be something like “my leg hurts, but my arm doesn’t”. In that case, we need to find the negation and assign it correctly.
All of this was the first step, but it was the most difficult one. Next, we analyze the slot filling. For example, we don’t know under what condition a patient’s arm hurts (or maybe it always hurts) and so we have to ask him about it.
We generate questions based on pre-written templates and question the person on all the items. Then, the patient can choose one of the suggested options or write a free-style answer.
This kind of questioning continues until all the slots for the identified entities are filled (or marked as missing/unknown). We then attempt to predict the diagnosis and formulate new questions to clarify it. This was done using recommendation systems, but I won’t go into detail since I didn’t do it.
When the convergence criterion is reached, the dialogue ends, and we give an anamnesis and a tentative diagnosis.
The key thing is the data
Working with data was very challenging in our project. In general, there are fewer texts in Russian than in English. But that’s half the trouble. We needed data in the medical domain. Unfortunately, there are almost no datasets of this kind for the Russian language. We were able to pull several million texts from open sources (such as forums), but there were several problems. These included:
- The style and content of texts from the forums are very different from what people will write to the chatbot;
- The data have not been labeled;
- There are no pre-trained NLP models on Russian-language medical texts. The closest thing is either English-language BioBERT or some Russian-language BERT. But in the first case, the language does not fit, and in the second case, the domain does not fit;
Thus, we had to:
- label the texts by ourselves;
- take into account the differences in the domain between the texts we have and those that we will get in production;
- train models from scratch or pre-train on our own data;
Data labeling was done in several stages:
In the beginning, we did it manually or using simple rule-based parsers and regex (I joined the project just at this stage). When I tried to train the NER model on these labels, I got the expected results - the model was often wrong, and in many cases, it made correct predictions on the samples with the wrong labels.
After a while, we decided it would be better to label properly. If we wanted to label something simple (like location names or people’s names), we wouldn’t have to worry about preparing the instruction because everyone would know what to label. But we needed to work with medical entities, so the first step was to make detailed instructions.
This turned out to be a lot harder than we initially expected. We tried different approaches to cover most cases: we labeled the texts individually, for example, and then got together to discuss controversial points. As a result, the instructions had the following information:
- the entity, its description, and what questions it answers;
- examples of cases;
- examples of misleading cases, examples of words that may seem relevant but aren’t;
I already wrote above that we knew the difference between our texts and those that could come to us in the prod. But by then, we already had a working version of the chatbot (even though almost without the ML part), so we could look at the logs and find out what people wrote. The results were frankly depressing: sometimes people described their illnesses very briefly, in one or two words; sometimes they listed almost all the diseases they had, not only what was bothering them right now; finally, many messages did not contain anything about health problems - people wrote that they were fine, joked (for example, complained that their “soul hurts”) or just wrote nonsense.
Based on all this, we decided to do the following:
- take short phrases (up to 50 words) for labeling and model training;
- add enough text without entities/classes to reduce the proportion of false positives;
- add augmentations;
As time passed, the labeled texts were piling up, but it quickly became clear to us that its quality left much to be desired. The point is that there was almost no control over the labels: each text was labeled by one person, and the quality of the labels was not validated. But I could not just say, “let’s do better,” I had to show that there were problems and propose solutions.
At that time, we had about 15,000 labeled texts. Among them, I found about 3,000 duplicates - identical or almost identical texts which had been labeled several times because, at the time, we did not do a preliminary check for duplicates. So the analysis of these labels revealed several problems:
- different people labeled NERs in the same texts differently: some labeled prepositions, some did not; some labeled “extra” words, some did not, and so on;
- sometimes, the same entity appears several times in the text. Some of the labelers highlight all such entities, some only the first;
- Finally, it also happened that in the text, one labeler labeled only one entity, and another labeled only the other.
This analysis was enough to change the labeling process. Finally, as a result of several iterations, we had the following procedure:
- First, we collected the data for the labeling. This was initially done by simple keyword searches, but later we implemented some kind of a simplified version of active learning: we collect a minimal dataset, train the model and then make predictions, calculate entropy and take the texts with the maximum entropy for labeling. And this wasn’t used directly but was combined with some other criteria in snorkel. This worked quite well;
- these texts were given to the labelers along with the instruction we prepared. They labeled the data in a customized Doccano. 5 people were working on each text;
- there was a special chat room in Telegram where the labelers could ask questions to the “controllers” (people who better understand how to do it) to clarify controversial points. This gave a considerable increase in the quality. Once, for the sake of an experiment, we tried to cancel this stage, and as a result, this iteration turned out to be much worse than usual;
- for some time, the resulting labels were pre-checked by validators. They would randomly take 10% of the labeled texts and check the quality; if it was better than 90%, the labels was sent to us; if not - the data was sent for re-labeling;
- we used a lot of postprocessing - it fixed many issues that were easier to do automatically than have people follow them. It removed extra prepositions and spaces, junk words, and more. This script was iteratively improved as we discovered new minor discrepancies in the labels. This, too, resulted in a significant increase in quality;
- then, we ran the script we had written to analyze the quality of the labels. It showed lots of information: the proportion of texts with full and partial label matches, labeled chunks of text (for the NER task), and examples of mismatched and matching texts. By the way, it was handy to look at examples of texts with the same labels because sometimes it happened that all labelers made the same mistake. This was a signal that we should update the instructions for them. The same is true for texts where no labeler highlighted anything;
- if the proportion of texts with a full label match was higher than 90%, we accepted this batch of texts, but if not, the texts were sent back;

We had numerous ideas to speed up labeling, but usually, it was a tradeoff between the labelers’ time, the data scientists’ time, and the models’ quality. Let’s look at some suggestions.
One idea was as follows: let’s say 1000 texts were labeled in the current iteration. Analysis of the labels showed that only 70% of the texts had a full cross-labeling match. So we suggested that not all 1000 texts should be sent for re-labeling in such cases, but only the 300 for which there was a discrepancy. This approach, of course, significantly accelerated the process, but at the cost of a slight deterioration in quality because in the 700 texts with matching labels, there will definitely be texts where all the labelers were wrong. And then, either data scientists have to look through all the texts and check them for errors, or we accept the deterioration of models because of the worsening of labels.
Another idea: we have a lot of entities (more than 50), and it would be great to label all entities in each text; in this case, it would be possible to train one model for all entities at once. Unfortunately, this was not possible. First, it is time-consuming: some entities frequently occur (more than half the time), and some are rare (less than 10 or even 5 percent). If you ask people to label all the entities in all the texts, they usually won’t label anything. And more importantly, if you ask someone to label 50 entities in a text, they will forget about many of them.
As a result, we just sent texts to the labelers and asked them to label one entity for a long time. Later we tried to give the same texts to label other entities or ask them to label up to five specific entities in the texts.
And even all of this was often lacking. For example, at one point, we found that in the phrase “my arm hurts,” the NER model for localization did not find the word “arm”. It turned out that out of four thousand texts labeled at the time, only one had the word “arm”.
Moreover, there were often complex cases in the texts where it was easy to make mistakes. Let me give you an example: “suffering from very severe headaches, sometimes even going dark in the eyes from the pain.” The word “headache” is the entity “pain”. But the word “pain” at the end is not the entity “pain” - instead, the condition in which the darkness in the eyes occurs.
More about machine learning
Text processing
Text processing caused us a lot of suffering.
Above I have already described the postprocessing for the standardization of data labels. We used similar postprocessing after entity extraction models. In addition, we had our own abbreviation decoding, postprocessing of predictions based on business rules, and much more.
Text tokenization deserves special attention. There are many tokenization methods, and it’s hard to say which is the best. Initially, we used the spaCy tokenizer (since spaCy was very actively used in the project), and switching to other tokenizers would have meant rewriting many pieces of the project, so we did not replace it. But it was often necessary to tweak it manually, so it didn’t break on our texts. An example of such code:
from spacy.tokenizer import Tokenizer
from spacy.util import compile_infix_regex, compile_suffix_regex, compile_prefix_regex
def custom_tokenizer(nlp):
"""Creates custom tokenizer for spacy"""
suf = list(nlp.Defaults.suffixes) # Default suffixes
# remove suffixes, so that spacy doesn't split things like '140мм рт ст'
del suf[75]
suffixes = compile_suffix_regex(tuple(suf))
# remove №
inf = list(nlp.Defaults.infixes)
inf[2] = inf[2].replace('\\u2116', '')
infix_re = compile_infix_regex(inf)
pre = list(nlp.Defaults.prefixes)
pre[-1] = pre[-1].replace('\\u2116', '')
pre_compiled = compile_prefix_regex(pre)
return Tokenizer(nlp.vocab,
prefix_search=pre_compiled.search,
suffix_search=suffixes.search,
infix_finditer=infix_re.finditer,
token_match=nlp.tokenizer.token_match,
rules=nlp.Defaults.tokenizer_exceptions)
Embeddings
In NLP, one of the keys to success is using good pre-trained models or at least pre-trained embeddings.
As I wrote above, there are no NLP models pre-trained on Russian medical texts, so we had to look for other approaches.
To begin with, I simply took the public embedding fasttext pre-trained on Wikipedia. They worked fine, but I wanted something better. So I took all our existing medical texts and began to train the embeddings on them - word2vec, glove, and fasttext. The fasttext embeddings turned out to be the best (which is not surprising), and the choice of hyperparameters also played an important role.
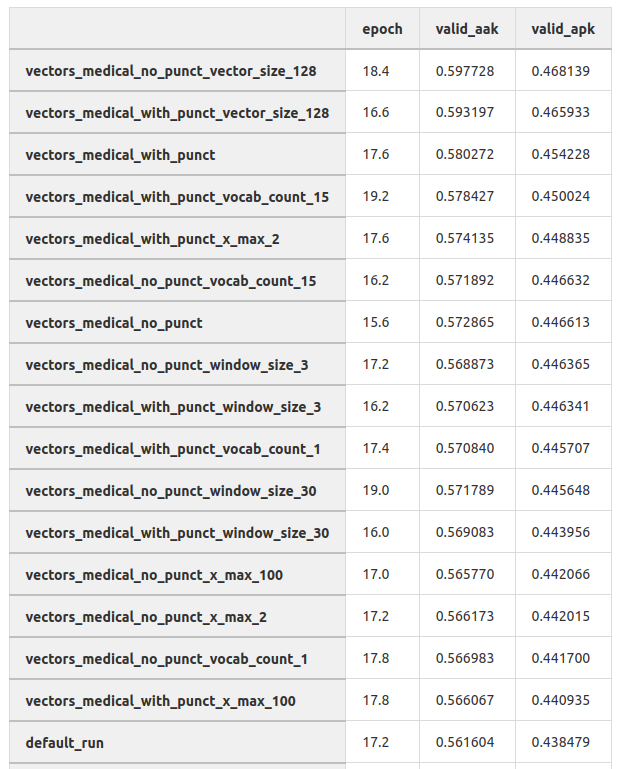
In this table, you can see the results of training the diagnosis classification model on different embeddings. This was an experiment on the direct prediction of diagnoses on the full text of the complaint. We did not use this approach, but nevertheless, we can see that selecting hyperparameters of the embedding can significantly increase the quality of the models.
NER models
Entity extraction started out simple: in the beginning, we used regex parsers to search the keywords. I want to note that this approach continued to be used for simple entities until the very end; we had entities easily extracted by keywords, so there was no need to spend resources on labeling them.
The next step was to use NER models from spaCy. That is, we either trained models from scratch or used spacy_ru. At that time, it was more convenient to train models using self-written python scripts, but in the new versions, it is much easier to do this simply in the command line. Finally, we tried combining training spacy models with EntityRuler - that is, the ability to add rules or just search by keywords, but it wasn’t very helpful.
When we accumulated more labels, we switched to neural nets. BiLSTM on fasttext vectors worked fine. We tried experimenting with architecture, for example adding attention, but it didn’t improve too much. In the end, we just tuned the architecture and hyperparameters of the model to suit different entities.
Classification
With classification, everything was straightforward: the models took very short texts as input - which is what the NER models predicted. It made no sense to train any complex models on this, so we just used the old, proven approach - vectorization with TF-IDF on letter combinations (char-grams) and word combinations (word-grams) and logistic regression for prediction.
It was convenient to use Pipeline from sklearn for this, and it looked something like this:
from sklearn.pipeline import FeatureUnion
from sklearn.feature_extraction.text import TfidfVectorizer
from sklearn.pipeline import Pipeline
from sklearn.linear_model import LogisticRegression
combined_features = FeatureUnion([('tfidf', TfidfVectorizer(ngram_range=(1, 3))),
('tfidf_char', TfidfVectorizer(ngram_range=(1, 3),
analyzer='char'))])
pipeline = Pipeline([('features', combined_features),
('clf', LogisticRegression(class_weight='balanced',
solver='lbfgs',
n_jobs=10,
multi_class='auto'))])
Relation extraction
Finding relationships between entities was quite a challenge. Sometimes the related entities were next to each other in the text; sometimes, they were far away; sometimes, there were one-to-one relationships in the texts; sometimes, many-to-many relationships; moreover, not all combinations of relationships were possible. All this made it challenging to train models because an inattentively assembled dataset easily led to overfitting and many false positives. We tried many models, e.g., we started by just extracting ELMo embeddings and MLPs on top of them, but this approach worked slowly and was not of much quality.
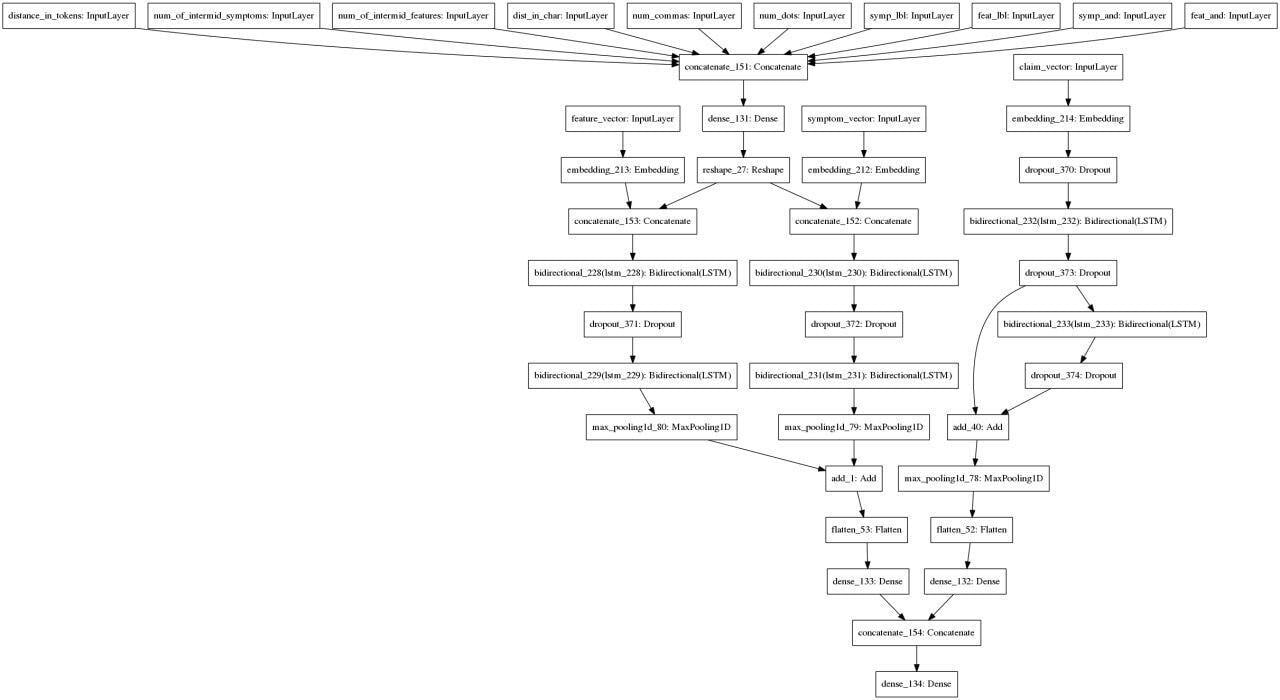
After many iterations, we came up with this approach: we take a sentence and entities extracted from it, vectorize it, extract additional attributes for each pair of entities and attributes and use it in this architecture:
Augmentation
Augmentations have played a pretty significant role in improving the quality of our models. The fact is that there were always not enough labels, and this has become one of our main problems. For example, the localization entity (recall, this is where the problem is - leg, arm, and so on) had more than 150 classes - which means that both NER models need to catch such words, and classification models need to correctly identify their classes, and relation extraction models need to accurately find links to such terms. The other entities had significantly fewer classes but still had similar difficulties. If we label texts randomly, there is a high probability that many classes/words will not be encountered. And searching for all classes manually is difficult. The attempt to manually collect texts with all the word pairs for the extraction models is doomed to fail.
We started using augmentations, most of them rule-based, rather than some clever variants. One standard text augmentation approach is replacing words with synonyms or words with similar embeddings. Unfortunately, this didn’t work in our case.
Let’s say there’s a phrase, “my arm hurts.” If we just try to take a synonym or word with a close embedding to the word “pain”, we might get something appropriate, like “soreness”, or we might get, for example, “discomfort”, “aching” - and these are already other entities. In addition, such substitutions may break the spelling.
One approach that has worked:
- take the original sentence, such as, again, “I have a pain in my arm” and generate new sentences by simply replacing “arm” with other localizations. We have to remember to use the correct inflections;
- further, we can change the words in the entity of “pain” and get something like “I have pain in my arm” or “my arm hurts” - and again, we have to check the forms of the words;
- in addition, we can add attributes and get “I have severe pain in my hand”, “I have pain in my hand in the morning,” and many other things. It is crucial to change the labels themselves as well;
- finally, we can replace the entity “pain” with something else and get “I have a pain in my arm”, “my arm is swollen”, and change the labels too;
And this approach can be used with both NER datasets and classification and extraction datasets.
All this sounds too good - as if the labels are not really needed. And indeed, it turned out to be too good to be true. This approach caused two problems: we didn’t know all the possible word options, and the models learned the patterns too quickly. As a result, we were getting wild overfits and many false positives.
To fix the situation, we had to randomize augmentations, add more junk words (not entities) to the generated phrases, and ensure that in the training dataset, a significant part of the texts was real, not generated. This approach proved to work.
Additional technical details
I’ll describe some other technical points that I found pretty interesting.
We had quite a few legacy scripts, most of which we kept, some we rewrote out of necessity, and some for convenience. For example, different classification/NER models were written in separate scripts, imported into another script, and used there. This worked fine, but when adding/modifying models, you would have to change the imports and make other changes to the code, which is not always a good thing. So I rewrote this with the result that model settings and class paths were stored in the YAML-config. Because of this, if we were training a new version of a model or adding a new model, we didn’t need to change the underlying code, just changed the config and, if necessary, added a script with the new model code.
single_entities_models:
localization:
model_name: pytorch_ner.PytorchNerWrapper
params:
path: localization.pth
ner_name: localization
idx2label_path: idx2label.json
config_path: config.json
preprocessing: True
productivity:
model_name: pytorch_ner.PytorchNerWrapper
params:
path: productivity.pth
ner_name: productivity
idx2label_path: productivity_pytorch_220920/idx2label.json
config_path: productivity_pytorch_220920/config.json
preprocessing: True
We set up a basic ci/cd, although it was more of a style check.
At some point, we decided that we needed to have more or less clear criteria on whether to roll out a new version of the model or not. To do this, we (data scientists) collected and labeled our own test dataset to check the quality of the models. It contained texts with full labels of entities, classes, and links between entities. Because of the labor-intensive nature of this process, there were only a few hundred examples. When we had a new version of a model, we would run it on this dataset and see how much the quality had changed, paying attention to both false positives and false negatives. The new model was accepted only if it improved all metrics. At the same time, this allowed us to report to our managers on the progress in improving the models.
One of the project’s challenges was that we had so many models - we had to extract 50+ entities, find connections between them, classify them, and so on. The result was a large total size of models and a slow project speed. For example, at one point, we simply couldn’t run it on a small server because there wasn’t enough RAM. Speed was also critical: the user must get a response quickly. We solved these difficulties by a combination of measures:
- Simply optimizing the code (in legacy code, it was not uncommon for the same model to be initialized many times or used inefficiently).
- Using models on rules where possible.
- Training one model on many entities if the labels allowed it.
It also meant that we couldn’t just cram a dozen BERTs into a project, which would have exceeded all possible limits.
The ideas that didn’t work
We had a lot of ideas that either didn’t work or just didn’t get to try for a number of reasons. I will list some of them.
We really wanted to train one model for all entities. And ideally, we wanted to make a complex architecture and train the model simultaneously for NER and Relation extraction (and, if possible, for classification as well). Unfortunately, this was limited by the availability of labeling, but we had no way to label a large enough dataset for all entities. However, we did manage to try SpERT, but the result was not good enough to implement it in the project.
We tried lemmatization but finally gave up on that idea. We have tested different tools: spacy, pymorphy2, natasha, rnnmorph, and others; pymorphy2 was the fastest and the best one on our data. But the use of lemmatization did not improve our models’ quality. In addition, many medical terms were not handled well by lemmatizers. Finally, the use of lemmatizers noticeably slowed down the system’s response speed, so we decided there was no point in using them.
The idea was to try using spellcheckers since many people write with mistakes. Our team tested Jamspell and pyenchant, but alas, they often corrupted the texts and slowed down the project considerably.
We also tried to convert trained models into other formats to speed up the inference, but it turned out that the CRF layer is not convertible to onnx, and if you train NER models without it, the quality drops significantly.
The reasons the project didn’t succeed
As mentioned at the beginning of this post, the project has been stopped. This decision was made for reasons beyond our control. Partly, it happened because the project was not working well enough. There are two groups of reasons: technical and organizational. However, they were often interrelated.
Technical problems
I’ve written a lot about working with data, and I’ll repeat it: labeling it was complicated and time-consuming. To improve the quality of the models and to cover different cases, it would have been worth spending much more time and effort on it.
We could not use the chatbot log data productively because there was very little of it. We should have done one of two things: either use the chatbot more actively and analyze the logs or focus project development on the areas where most of the complaints were received.
Organizational problems
- Most of the time, we either had no roadmap, or it was very top-level. Because of this, it was not very clear what had to be done to make the project successful;
- Because of this, from time to time, new ideas came up from product managers, leading to changes in functionality, changes in the list of retrievable entities and classes, and changes in the logic of work. Often the ideas were ill-conceived, and we had to tweak them ourselves;
- What’s more, we had no clear-cut criteria for the quality of our models. We spent a lot of time and effort on improving our models, but we had no understanding of what quality of models was sufficient. We even assembled a test dataset on our own to check the quality of our models;
- In addition, we did not have any metrics to assess the quality of the dialogue. That is, we could measure the quality of the models with the usual machine learning metrics, but we had no way to assess whether our dialog system as a whole was working well or not;
- We had no testing. The project was very complex; in addition to the NLP part, there was a large part responsible for the dialogue itself, an impressive backend component, etc. We found some bugs and fixed them from time to time, but it would have been much better if we had a QA team. To be fair, we could hardly ask QA to test the project from a medical point of view because that would require domain knowledge, but even beyond that, there were many things to test;
Somewhere near the end of the project, three business metrics were announced to us:
- the person agreed to the recommendation of the chatbot;
- the person agreed and made an appointment;
- the person agreed, made an appointment, and came in for an appointment.
The problem was that we could only influence the first metric; the second and third metrics did not depend on the quality of the chatbot.
As a result, the project was very complicated, with no clear understanding of how well it would work in real conditions and how much money it would bring. As a result, it was frozen by a decision from above at the end of 2020.
Was there any benefit from the project?
Well, what was the result? It would seem all for nothing: many resources were spent, and the project was stopped. But nevertheless, when asked if there was any benefit to it, I would say yes. And here’s why:
- We tried many approaches to active learning and analysis of label quality, and some of these approaches were used in future projects;
- We gained experience building different models for working with texts - NER, classification, and relation extraction. This was also used in further projects;
- While training models, we developed two pipelines on PyTorch lightning - these pipelines can be used again in the future;
The project is frozen but may be resumed in the future.
That’s the whole story of this project. I hope it was interesting and valuable. :)
blogpost nlp ner relationextraction classification